2 DataFrame
DataFrame is a 2-dimensional labeled data structure with columns of potentially different types. You can think of it like a spreadsheet or SQL table, or a dict of Series objects. It is generally the most commonly used pandas object. Like Series, DataFrame accepts many different kinds of input:
- Dict of 1D ndarrays, lists, dicts, or Series
- 2-D numpy.ndarray
- Structured or record ndarray
- A
Series- Another
DataFrame
Along with the data, you can optionally pass index (row labels) and columns (column labels) arguments. If you pass an index and / or columns, you are guaranteeing the index and / or columns of the resulting DataFrame. Thus, a dict of Series plus a specific index will discard all data not matching up to the passed index.
If axis labels are not passed, they will be constructed from the input data based on common sense rules.
2.1 From dict of Series or dicts
The result index will be the union of the indexes of the various Series. If there are any nested dicts, these will be first converted to Series. If no columns are passed, the columns will be the sorted list of dict keys.
In [1]: d = {'one' : pd.Series([1., 2., 3.], index=['a', 'b', 'c']),
...: 'two' : pd.Series([1., 2., 3., 4.], index=['a', 'b', 'c', 'd'])}
...:
In [2]: df = pd.DataFrame(d)
In [3]: df
Out[3]:
one two
a 1.0 1.0
b 2.0 2.0
c 3.0 3.0
d NaN 4.0
In [4]: pd.DataFrame(d, index=['d', 'b', 'a'])
Out[4]:
one two
d NaN 4.0
b 2.0 2.0
a 1.0 1.0
In [5]: pd.DataFrame(d, index=['d', 'b', 'a'], columns=['two', 'three'])
Out[5]:
two three
d 4.0 NaN
b 2.0 NaN
a 1.0 NaN
The row and column labels can be accessed respectively by accessing the index and columns attributes:
Note
When a particular set of columns is passed along with a dict of data, the passed columns override the keys in the dict.
In [6]: df.index
Out[6]: Index([u'a', u'b', u'c', u'd'], dtype='object')
In [7]: df.columns
Out[7]: Index([u'one', u'two'], dtype='object')
2.2 From dict of ndarrays / lists
The ndarrays must all be the same length. If an index is passed, it must
clearly also be the same length as the arrays. If no index is passed, the
result will be range(n), where n is the array length.
In [8]: d = {'one' : [1., 2., 3., 4.],
...: 'two' : [4., 3., 2., 1.]}
...:
In [9]: pd.DataFrame(d)
Out[9]:
one two
0 1.0 4.0
1 2.0 3.0
2 3.0 2.0
3 4.0 1.0
In [10]: pd.DataFrame(d, index=['a', 'b', 'c', 'd'])
Out[10]:
one two
a 1.0 4.0
b 2.0 3.0
c 3.0 2.0
d 4.0 1.0
2.3 From structured or record array
This case is handled identically to a dict of arrays.
In [11]: data = np.zeros((2,), dtype=[('A', 'i4'),('B', 'f4'),('C', 'a10')])
In [12]: data[:] = [(1,2.,'Hello'), (2,3.,"World")]
In [13]: pd.DataFrame(data)
Out[13]:
A B C
0 1 2.0 Hello
1 2 3.0 World
In [14]: pd.DataFrame(data, index=['first', 'second'])
Out[14]:
A B C
first 1 2.0 Hello
second 2 3.0 World
In [15]: pd.DataFrame(data, columns=['C', 'A', 'B'])
Out[15]:
C A B
0 Hello 1 2.0
1 World 2 3.0
Note
DataFrame is not intended to work exactly like a 2-dimensional NumPy ndarray.
2.4 From a list of dicts
In [16]: data2 = [{'a': 1, 'b': 2}, {'a': 5, 'b': 10, 'c': 20}]
In [17]: pd.DataFrame(data2)
Out[17]:
a b c
0 1 2 NaN
1 5 10 20.0
In [18]: pd.DataFrame(data2, index=['first', 'second'])
Out[18]:
a b c
first 1 2 NaN
second 5 10 20.0
In [19]: pd.DataFrame(data2, columns=['a', 'b'])
Out[19]:
a b
0 1 2
1 5 10
2.5 From a dict of tuples
You can automatically create a multi-indexed frame by passing a tuples dictionary
In [20]: pd.DataFrame({('a', 'b'): {('A', 'B'): 1, ('A', 'C'): 2},
....: ('a', 'a'): {('A', 'C'): 3, ('A', 'B'): 4},
....: ('a', 'c'): {('A', 'B'): 5, ('A', 'C'): 6},
....: ('b', 'a'): {('A', 'C'): 7, ('A', 'B'): 8},
....: ('b', 'b'): {('A', 'D'): 9, ('A', 'B'): 10}})
....:
Out[20]:
a b
a b c a b
A B 4.0 1.0 5.0 8.0 10.0
C 3.0 2.0 6.0 7.0 NaN
D NaN NaN NaN NaN 9.0
2.6 From a Series
The result will be a DataFrame with the same index as the input Series, and with one column whose name is the original name of the Series (only if no other column name provided).
Missing Data
Much more will be said on this topic in the Missing data
section. To construct a DataFrame with missing data, use np.nan for those
values which are missing. Alternatively, you may pass a numpy.MaskedArray
as the data argument to the DataFrame constructor, and its masked entries will
be considered missing.
2.7 Alternate Constructors
DataFrame.from_dict
DataFrame.from_dict takes a dict of dicts or a dict of array-like sequences
and returns a DataFrame. It operates like the DataFrame constructor except
for the orient parameter which is 'columns' by default, but which can be
set to 'index' in order to use the dict keys as row labels.
DataFrame.from_records
DataFrame.from_records takes a list of tuples or an ndarray with structured
dtype. Works analogously to the normal DataFrame constructor, except that
index maybe be a specific field of the structured dtype to use as the index.
For example:
In [21]: data
Out[21]:
array([(1, 2.0, 'Hello'), (2, 3.0, 'World')],
dtype=[('A', '<i4'), ('B', '<f4'), ('C', 'S10')])
In [22]: pd.DataFrame.from_records(data, index='C')
Out[22]:
A B
C
Hello 1 2.0
World 2 3.0
DataFrame.from_items
DataFrame.from_items works analogously to the form of the dict
constructor that takes a sequence of (key, value) pairs, where the keys are
column (or row, in the case of orient='index') names, and the value are the
column values (or row values). This can be useful for constructing a DataFrame
with the columns in a particular order without having to pass an explicit list
of columns:
In [23]: pd.DataFrame.from_items([('A', [1, 2, 3]), ('B', [4, 5, 6])])
Out[23]:
A B
0 1 4
1 2 5
2 3 6
If you pass orient='index', the keys will be the row labels. But in this
case you must also pass the desired column names:
In [24]: pd.DataFrame.from_items([('A', [1, 2, 3]), ('B', [4, 5, 6])],
....: orient='index', columns=['one', 'two', 'three'])
....:
Out[24]:
one two three
A 1 2 3
B 4 5 6
2.8 Column selection, addition, deletion
You can treat a DataFrame semantically like a dict of like-indexed Series objects. Getting, setting, and deleting columns works with the same syntax as the analogous dict operations:
In [25]: df['one']
Out[25]:
a 1.0
b 2.0
c 3.0
d NaN
Name: one, dtype: float64
In [26]: df['three'] = df['one'] * df['two']
In [27]: df['flag'] = df['one'] > 2
In [28]: df
Out[28]:
one two three flag
a 1.0 1.0 1.0 False
b 2.0 2.0 4.0 False
c 3.0 3.0 9.0 True
d NaN 4.0 NaN False
Columns can be deleted or popped like with a dict:
In [29]: del df['two']
In [30]: three = df.pop('three')
In [31]: df
Out[31]:
one flag
a 1.0 False
b 2.0 False
c 3.0 True
d NaN False
When inserting a scalar value, it will naturally be propagated to fill the column:
In [32]: df['foo'] = 'bar'
In [33]: df
Out[33]:
one flag foo
a 1.0 False bar
b 2.0 False bar
c 3.0 True bar
d NaN False bar
When inserting a Series that does not have the same index as the DataFrame, it will be conformed to the DataFrame’s index:
In [34]: df['one_trunc'] = df['one'][:2]
In [35]: df
Out[35]:
one flag foo one_trunc
a 1.0 False bar 1.0
b 2.0 False bar 2.0
c 3.0 True bar NaN
d NaN False bar NaN
You can insert raw ndarrays but their length must match the length of the DataFrame’s index.
By default, columns get inserted at the end. The insert function is
available to insert at a particular location in the columns:
In [36]: df.insert(1, 'bar', df['one'])
In [37]: df
Out[37]:
one bar flag foo one_trunc
a 1.0 1.0 False bar 1.0
b 2.0 2.0 False bar 2.0
c 3.0 3.0 True bar NaN
d NaN NaN False bar NaN
2.9 Assigning New Columns in Method Chains
New in version 0.16.0.
Inspired by dplyr’s
mutate verb, DataFrame has an assign()
method that allows you to easily create new columns that are potentially
derived from existing columns.
In [38]: iris = pd.read_csv('https://raw.githubusercontent.com/pydata/pandas/master/doc/data/iris.data')
In [39]: iris.head()
Out[39]:
SepalLength SepalWidth PetalLength PetalWidth Name
0 5.1 3.5 1.4 0.2 Iris-setosa
1 4.9 3.0 1.4 0.2 Iris-setosa
2 4.7 3.2 1.3 0.2 Iris-setosa
3 4.6 3.1 1.5 0.2 Iris-setosa
4 5.0 3.6 1.4 0.2 Iris-setosa
In [40]: (iris.assign(sepal_ratio = iris['SepalWidth'] / iris['SepalLength'])
....: .head())
....:
Out[40]:
SepalLength SepalWidth PetalLength PetalWidth Name sepal_ratio
0 5.1 3.5 1.4 0.2 Iris-setosa 0.6863
1 4.9 3.0 1.4 0.2 Iris-setosa 0.6122
2 4.7 3.2 1.3 0.2 Iris-setosa 0.6809
3 4.6 3.1 1.5 0.2 Iris-setosa 0.6739
4 5.0 3.6 1.4 0.2 Iris-setosa 0.7200
Above was an example of inserting a precomputed value. We can also pass in a function of one argument to be evalutated on the DataFrame being assigned to.
In [41]: iris.assign(sepal_ratio = lambda x: (x['SepalWidth'] /
....: x['SepalLength'])).head()
....:
Out[41]:
SepalLength SepalWidth PetalLength PetalWidth Name sepal_ratio
0 5.1 3.5 1.4 0.2 Iris-setosa 0.6863
1 4.9 3.0 1.4 0.2 Iris-setosa 0.6122
2 4.7 3.2 1.3 0.2 Iris-setosa 0.6809
3 4.6 3.1 1.5 0.2 Iris-setosa 0.6739
4 5.0 3.6 1.4 0.2 Iris-setosa 0.7200
assign always returns a copy of the data, leaving the original
DataFrame untouched.
Passing a callable, as opposed to an actual value to be inserted, is
useful when you don’t have a reference to the DataFrame at hand. This is
common when using assign in chains of operations. For example,
we can limit the DataFrame to just those observations with a Sepal Length
greater than 5, calculate the ratio, and plot:
In [42]: (iris.query('SepalLength > 5')
....: .assign(SepalRatio = lambda x: x.SepalWidth / x.SepalLength,
....: PetalRatio = lambda x: x.PetalWidth / x.PetalLength)
....: .plot(kind='scatter', x='SepalRatio', y='PetalRatio'))
....:
Out[42]: <matplotlib.axes._subplots.AxesSubplot at 0x2b35ba0c5cd0>
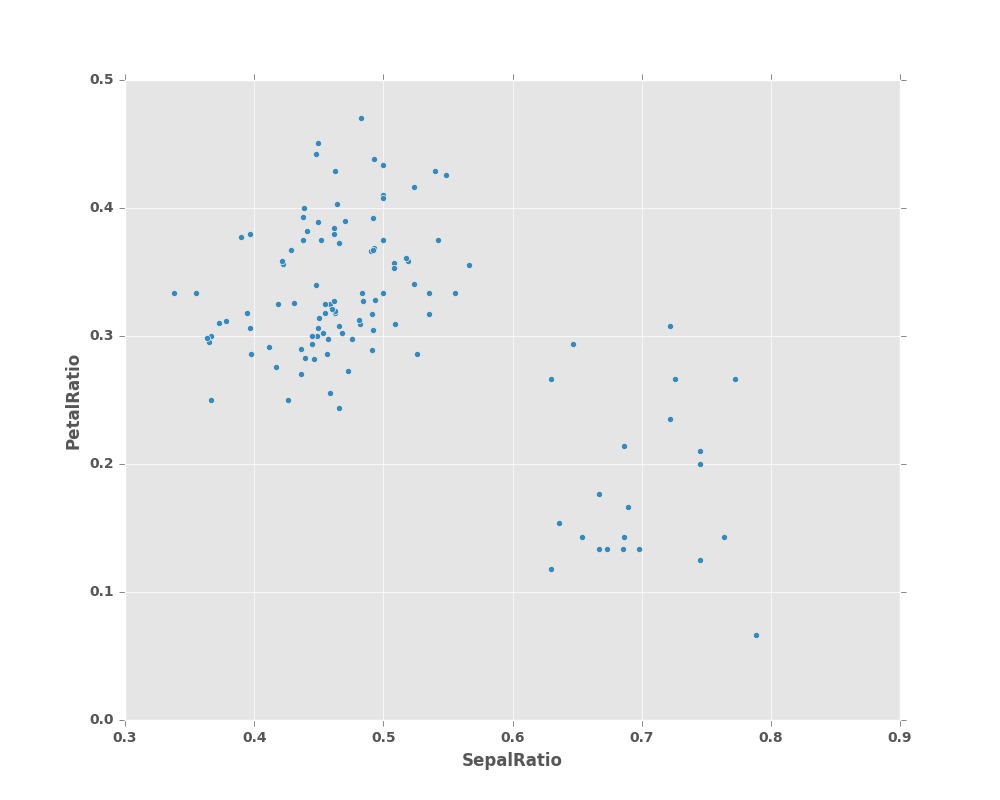
Since a function is passed in, the function is computed on the DataFrame being assigned to. Importantly, this is the DataFrame that’s been filtered to those rows with sepal length greater than 5. The filtering happens first, and then the ratio calculations. This is an example where we didn’t have a reference to the filtered DataFrame available.
The function signature for assign is simply **kwargs. The keys
are the column names for the new fields, and the values are either a value
to be inserted (for example, a Series or NumPy array), or a function
of one argument to be called on the DataFrame. A copy of the original
DataFrame is returned, with the new values inserted.
Warning
Since the function signature of assign is **kwargs, a dictionary,
the order of the new columns in the resulting DataFrame cannot be guaranteed
to match the order you pass in. To make things predictable, items are inserted
alphabetically (by key) at the end of the DataFrame.
All expressions are computed first, and then assigned. So you can’t refer
to another column being assigned in the same call to assign. For example:
In [43]: # Don't do this, bad reference to `C` df.assign(C = lambda x: x['A'] + x['B'], D = lambda x: x['A'] + x['C']) In [2]: # Instead, break it into two assigns (df.assign(C = lambda x: x['A'] + x['B']) .assign(D = lambda x: x['A'] + x['C']))
2.10 Indexing / Selection
The basics of indexing are as follows:
| Operation | Syntax | Result |
|---|---|---|
| Select column | df[col] |
Series |
| Select row by label | df.loc[label] |
Series |
| Select row by integer location | df.iloc[loc] |
Series |
| Slice rows | df[5:10] |
DataFrame |
| Select rows by boolean vector | df[bool_vec] |
DataFrame |
Row selection, for example, returns a Series whose index is the columns of the DataFrame:
In [44]: df.loc['b']
Out[44]:
one 2
bar 2
flag False
foo bar
one_trunc 2
Name: b, dtype: object
In [45]: df.iloc[2]
Out[45]:
one 3
bar 3
flag True
foo bar
one_trunc NaN
Name: c, dtype: object
For a more exhaustive treatment of more sophisticated label-based indexing and slicing, see the section on indexing. We will address the fundamentals of reindexing / conforming to new sets of labels in the section on reindexing.
2.11 Data alignment and arithmetic
Data alignment between DataFrame objects automatically align on both the columns and the index (row labels). Again, the resulting object will have the union of the column and row labels.
In [46]: df = pd.DataFrame(np.random.randn(10, 4), columns=['A', 'B', 'C', 'D'])
In [47]: df2 = pd.DataFrame(np.random.randn(7, 3), columns=['A', 'B', 'C'])
In [48]: df + df2
Out[48]:
A B C D
0 -0.8698 0.0952 -3.0582 NaN
1 -2.0222 0.2924 0.1788 NaN
2 0.6818 -0.0208 0.9536 NaN
3 -0.2146 0.1853 -1.0567 NaN
.. ... ... ... ..
6 -2.2563 -2.5575 0.1759 NaN
7 NaN NaN NaN NaN
8 NaN NaN NaN NaN
9 NaN NaN NaN NaN
[10 rows x 4 columns]
When doing an operation between DataFrame and Series, the default behavior is to align the Series index on the DataFrame columns, thus broadcasting row-wise. For example:
In [49]: df - df.iloc[0]
Out[49]:
A B C D
0 0.0000 0.0000 0.0000 0.0000
1 -1.6515 1.0883 2.0536 0.4969
2 -0.3710 0.3121 1.7708 0.0459
3 0.5387 1.4798 0.5705 3.5346
.. ... ... ... ...
6 -0.4335 -0.5131 2.9070 -0.4678
7 -1.7707 2.2825 2.8011 1.5385
8 0.5189 -1.5368 2.7514 -1.0378
9 -0.7670 0.5960 2.3229 -0.2503
[10 rows x 4 columns]
In the special case of working with time series data, and the DataFrame index also contains dates, the broadcasting will be column-wise:
In [50]: index = pd.date_range('1/1/2000', periods=8)
In [51]: df = pd.DataFrame(np.random.randn(8, 3), index=index, columns=list('ABC'))
In [52]: df
Out[52]:
A B C
2000-01-01 1.4627 -1.7432 -0.8266
2000-01-02 -0.3454 1.3142 0.6906
2000-01-03 0.9958 2.3968 0.0149
2000-01-04 3.3574 -0.3174 -1.2363
2000-01-05 0.8962 -0.4876 -0.0822
2000-01-06 -2.1829 0.3804 0.0848
2000-01-07 0.4324 1.5200 -0.4937
2000-01-08 0.6002 0.2742 0.1329
In [53]: type(df['A'])
Out[53]: pandas.core.series.Series
In [54]: df - df['A']
Out[54]:
2000-01-01 00:00:00 2000-01-02 00:00:00 2000-01-03 00:00:00 \
2000-01-01 NaN NaN NaN
2000-01-02 NaN NaN NaN
2000-01-03 NaN NaN NaN
2000-01-04 NaN NaN NaN
2000-01-05 NaN NaN NaN
2000-01-06 NaN NaN NaN
2000-01-07 NaN NaN NaN
2000-01-08 NaN NaN NaN
2000-01-04 00:00:00 ... 2000-01-08 00:00:00 A B C
2000-01-01 NaN ... NaN NaN NaN NaN
2000-01-02 NaN ... NaN NaN NaN NaN
2000-01-03 NaN ... NaN NaN NaN NaN
2000-01-04 NaN ... NaN NaN NaN NaN
2000-01-05 NaN ... NaN NaN NaN NaN
2000-01-06 NaN ... NaN NaN NaN NaN
2000-01-07 NaN ... NaN NaN NaN NaN
2000-01-08 NaN ... NaN NaN NaN NaN
[8 rows x 11 columns]
Warning
df - df['A']
is now deprecated and will be removed in a future release. The preferred way to replicate this behavior is
df.sub(df['A'], axis=0)
For explicit control over the matching and broadcasting behavior, see the section on flexible binary operations.
Operations with scalars are just as you would expect:
In [55]: df * 5 + 2
Out[55]:
A B C
2000-01-01 9.3135 -6.7158 -2.1330
2000-01-02 0.2732 8.5712 5.4529
2000-01-03 6.9788 13.9839 2.0744
2000-01-04 18.7871 0.4128 -4.1813
2000-01-05 6.4809 -0.4380 1.5888
2000-01-06 -8.9147 3.9020 2.4242
2000-01-07 4.1619 9.5999 -0.4683
2000-01-08 5.0009 3.3711 2.6644
In [56]: 1 / df
Out[56]:
A B C
2000-01-01 0.6837 -0.5737 -1.2098
2000-01-02 -2.8956 0.7609 1.4481
2000-01-03 1.0043 0.4172 67.2452
2000-01-04 0.2978 -3.1502 -0.8089
2000-01-05 1.1159 -2.0509 -12.1595
2000-01-06 -0.4581 2.6288 11.7863
2000-01-07 2.3127 0.6579 -2.0257
2000-01-08 1.6662 3.6466 7.5253
In [57]: df ** 4
Out[57]:
A B C
2000-01-01 4.5774 9.2332 4.6683e-01
2000-01-02 0.0142 2.9832 2.2743e-01
2000-01-03 0.9832 32.9999 4.8905e-08
2000-01-04 127.0651 0.0102 2.3359e+00
2000-01-05 0.6450 0.0565 4.5745e-05
2000-01-06 22.7073 0.0209 5.1819e-05
2000-01-07 0.0350 5.3375 5.9391e-02
2000-01-08 0.1298 0.0057 3.1182e-04
Boolean operators work as well:
In [58]: df1 = pd.DataFrame({'a' : [1, 0, 1], 'b' : [0, 1, 1] }, dtype=bool)
In [59]: df2 = pd.DataFrame({'a' : [0, 1, 1], 'b' : [1, 1, 0] }, dtype=bool)
In [60]: df1 & df2
Out[60]:
a b
0 False False
1 False True
2 True False
In [61]: df1 | df2
Out[61]:
a b
0 True True
1 True True
2 True True
In [62]: df1 ^ df2
Out[62]:
a b
0 True True
1 True False
2 False True
In [63]: -df1
Out[63]:
a b
0 False True
1 True False
2 False False
2.12 Transposing
To transpose, access the T attribute (also the transpose function),
similar to an ndarray:
# only show the first 5 rows
In [64]: df[:5].T
Out[64]:
2000-01-01 2000-01-02 2000-01-03 2000-01-04 2000-01-05
A 1.4627 -0.3454 0.9958 3.3574 0.8962
B -1.7432 1.3142 2.3968 -0.3174 -0.4876
C -0.8266 0.6906 0.0149 -1.2363 -0.0822
2.13 DataFrame interoperability with NumPy functions
Elementwise NumPy ufuncs (log, exp, sqrt, ...) and various other NumPy functions can be used with no issues on DataFrame, assuming the data within are numeric:
In [65]: np.exp(df)
Out[65]:
A B C
2000-01-01 4.3176 0.1750 0.4375
2000-01-02 0.7080 3.7219 1.9949
2000-01-03 2.7068 10.9877 1.0150
2000-01-04 28.7152 0.7280 0.2905
2000-01-05 2.4502 0.6141 0.9211
2000-01-06 0.1127 1.4629 1.0885
2000-01-07 1.5409 4.5721 0.6104
2000-01-08 1.8224 1.3155 1.1421
In [66]: np.asarray(df)
Out[66]:
array([[ 1.4627, -1.7432, -0.8266],
[-0.3454, 1.3142, 0.6906],
[ 0.9958, 2.3968, 0.0149],
[ 3.3574, -0.3174, -1.2363],
[ 0.8962, -0.4876, -0.0822],
[-2.1829, 0.3804, 0.0848],
[ 0.4324, 1.52 , -0.4937],
[ 0.6002, 0.2742, 0.1329]])
The dot method on DataFrame implements matrix multiplication:
In [67]: df.T.dot(df)
Out[67]:
A B C
A 20.6381 -2.1283 -5.9760
B -2.1283 13.3791 2.1350
C -5.9760 2.1350 2.9641
Similarly, the dot method on Series implements dot product:
In [68]: s1 = pd.Series(np.arange(5,10))
In [69]: s1.dot(s1)
Out[69]: 255
DataFrame is not intended to be a drop-in replacement for ndarray as its indexing semantics are quite different in places from a matrix.
2.14 Console display
Very large DataFrames will be truncated to display them in the console.
You can also get a summary using info().
(Here I am reading a CSV version of the baseball dataset from the plyr
R package):
In [70]: baseball = pd.read_csv('https://raw.githubusercontent.com/pydata/pandas/master/doc/data/baseball.csv')
In [71]: print(baseball)
id player year stint ... hbp sh sf gidp
0 88641 womacto01 2006 2 ... 0.0 3.0 0.0 0.0
1 88643 schilcu01 2006 1 ... 0.0 0.0 0.0 0.0
.. ... ... ... ... ... ... ... ... ...
98 89533 aloumo01 2007 1 ... 2.0 0.0 3.0 13.0
99 89534 alomasa02 2007 1 ... 0.0 0.0 0.0 0.0
[100 rows x 23 columns]
In [72]: baseball.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 100 entries, 0 to 99
Data columns (total 23 columns):
id 100 non-null int64
player 100 non-null object
year 100 non-null int64
stint 100 non-null int64
team 100 non-null object
lg 100 non-null object
g 100 non-null int64
ab 100 non-null int64
r 100 non-null int64
h 100 non-null int64
X2b 100 non-null int64
X3b 100 non-null int64
hr 100 non-null int64
rbi 100 non-null float64
sb 100 non-null float64
cs 100 non-null float64
bb 100 non-null int64
so 100 non-null float64
ibb 100 non-null float64
hbp 100 non-null float64
sh 100 non-null float64
sf 100 non-null float64
gidp 100 non-null float64
dtypes: float64(9), int64(11), object(3)
memory usage: 18.0+ KB
However, using to_string will return a string representation of the
DataFrame in tabular form, though it won’t always fit the console width:
In [73]: print(baseball.iloc[-20:, :12].to_string())
id player year stint team lg g ab r h X2b X3b
80 89474 finlest01 2007 1 COL NL 43 94 9 17 3 0
81 89480 embreal01 2007 1 OAK AL 4 0 0 0 0 0
82 89481 edmonji01 2007 1 SLN NL 117 365 39 92 15 2
83 89482 easleda01 2007 1 NYN NL 76 193 24 54 6 0
84 89489 delgaca01 2007 1 NYN NL 139 538 71 139 30 0
85 89493 cormirh01 2007 1 CIN NL 6 0 0 0 0 0
86 89494 coninje01 2007 2 NYN NL 21 41 2 8 2 0
87 89495 coninje01 2007 1 CIN NL 80 215 23 57 11 1
88 89497 clemero02 2007 1 NYA AL 2 2 0 1 0 0
89 89498 claytro01 2007 2 BOS AL 8 6 1 0 0 0
90 89499 claytro01 2007 1 TOR AL 69 189 23 48 14 0
91 89501 cirilje01 2007 2 ARI NL 28 40 6 8 4 0
92 89502 cirilje01 2007 1 MIN AL 50 153 18 40 9 2
93 89521 bondsba01 2007 1 SFN NL 126 340 75 94 14 0
94 89523 biggicr01 2007 1 HOU NL 141 517 68 130 31 3
95 89525 benitar01 2007 2 FLO NL 34 0 0 0 0 0
96 89526 benitar01 2007 1 SFN NL 19 0 0 0 0 0
97 89530 ausmubr01 2007 1 HOU NL 117 349 38 82 16 3
98 89533 aloumo01 2007 1 NYN NL 87 328 51 112 19 1
99 89534 alomasa02 2007 1 NYN NL 8 22 1 3 1 0
New since 0.10.0, wide DataFrames will now be printed across multiple rows by default:
In [74]: pd.DataFrame(np.random.randn(3, 12))
Out[74]:
0 1 2 3 4 5 6 \
0 -0.023688 2.410179 1.450520 0.206053 -0.251905 -2.213588 1.063327
1 -0.025747 -0.988387 0.094055 1.262731 1.289997 0.082423 -0.055758
2 -0.281461 0.030711 0.109121 1.126203 -0.977349 1.474071 -0.064034
7 8 9 10 11
0 1.266143 0.299368 -0.863838 0.408204 -1.048089
1 0.536580 -0.489682 0.369374 -0.034571 -2.484478
2 -1.282782 0.781836 -1.071357 0.441153 2.353925
You can change how much to print on a single row by setting the display.width
option:
In [75]: pd.set_option('display.width', 40) # default is 80
In [76]: pd.DataFrame(np.random.randn(3, 12))
Out[76]:
0 1 2 \
0 0.583787 0.221471 -0.744471
1 0.888782 0.228440 0.901805
2 1.574159 1.588931 0.476720
3 4 5 \
0 0.758527 1.729689 -0.964980
1 1.171216 0.520260 -1.197071
2 0.473424 -0.242861 -0.014805
6 7 8 \
0 -0.845696 -1.340896 1.846883
1 -1.066969 -0.303421 -0.858447
2 -0.284319 0.650776 -1.461665
9 10 11
0 -1.328865 1.682706 -1.717693
1 0.306996 -0.028665 0.384316
2 -1.137707 -0.891060 -0.693921
You can adjust the max width of the individual columns by setting display.max_colwidth
In [77]: datafile={'filename': ['filename_01','filename_02'],
....: 'path': ["media/user_name/storage/folder_01/filename_01",
....: "media/user_name/storage/folder_02/filename_02"]}
....:
In [78]: pd.set_option('display.max_colwidth',30)
In [79]: pd.DataFrame(datafile)
Out[79]:
filename \
0 filename_01
1 filename_02
path
0 media/user_name/storage/fo...
1 media/user_name/storage/fo...
In [80]: pd.set_option('display.max_colwidth',100)
In [81]: pd.DataFrame(datafile)
Out[81]:
filename \
0 filename_01
1 filename_02
path
0 media/user_name/storage/folder_01/filename_01
1 media/user_name/storage/folder_02/filename_02
You can also disable this feature via the expand_frame_repr option.
This will print the table in one block.
2.15 DataFrame column attribute access and IPython completion
If a DataFrame column label is a valid Python variable name, the column can be accessed like attributes:
In [82]: df = pd.DataFrame({'foo1' : np.random.randn(5),
....: 'foo2' : np.random.randn(5)})
....:
In [83]: df
Out[83]:
foo1 foo2
0 1.613616 -2.290613
1 0.464000 -1.134623
2 0.227371 -1.561819
3 -0.496922 -0.260838
4 0.306389 0.281957
In [84]: df.foo1
Out[84]:
0 1.613616
1 0.464000
2 0.227371
3 -0.496922
4 0.306389
Name: foo1, dtype: float64
The columns are also connected to the IPython completion mechanism so they can be tab-completed:
In [5]: df.fo<TAB>
df.foo1 df.foo2