5.6 Transformation
The transform method returns an object that is indexed the same (same size)
as the one being grouped. Thus, the passed transform function should return a
result that is the same size as the group chunk. For example, suppose we wished
to standardize the data within each group:
In [1]: index = pd.date_range('10/1/1999', periods=1100)
In [2]: ts = pd.Series(np.random.normal(0.5, 2, 1100), index)
In [3]: ts = ts.rolling(window=100,min_periods=100).mean().dropna()
In [4]: ts.head()
Out[4]:
2000-01-08 0.073603
2000-01-09 0.057313
2000-01-10 0.089255
2000-01-11 0.133248
2000-01-12 0.175876
Freq: D, dtype: float64
In [5]: ts.tail()
Out[5]:
2002-09-30 0.858589
2002-10-01 0.863873
2002-10-02 0.885344
2002-10-03 0.878466
2002-10-04 0.919510
Freq: D, dtype: float64
In [6]: key = lambda x: x.year
In [7]: zscore = lambda x: (x - x.mean()) / x.std()
In [8]: transformed = ts.groupby(key).transform(zscore)
We would expect the result to now have mean 0 and standard deviation 1 within each group, which we can easily check:
# Original Data
In [9]: grouped = ts.groupby(key)
In [10]: grouped.mean()
Out[10]:
2000 0.407865
2001 0.358106
2002 0.619029
dtype: float64
In [11]: grouped.std()
Out[11]:
2000 0.180659
2001 0.227780
2002 0.296282
dtype: float64
# Transformed Data
In [12]: grouped_trans = transformed.groupby(key)
In [13]: grouped_trans.mean()
Out[13]:
2000 -1.662242e-17
2001 7.592100e-16
2002 -4.344699e-16
dtype: float64
In [14]: grouped_trans.std()
Out[14]:
2000 1.0
2001 1.0
2002 1.0
dtype: float64
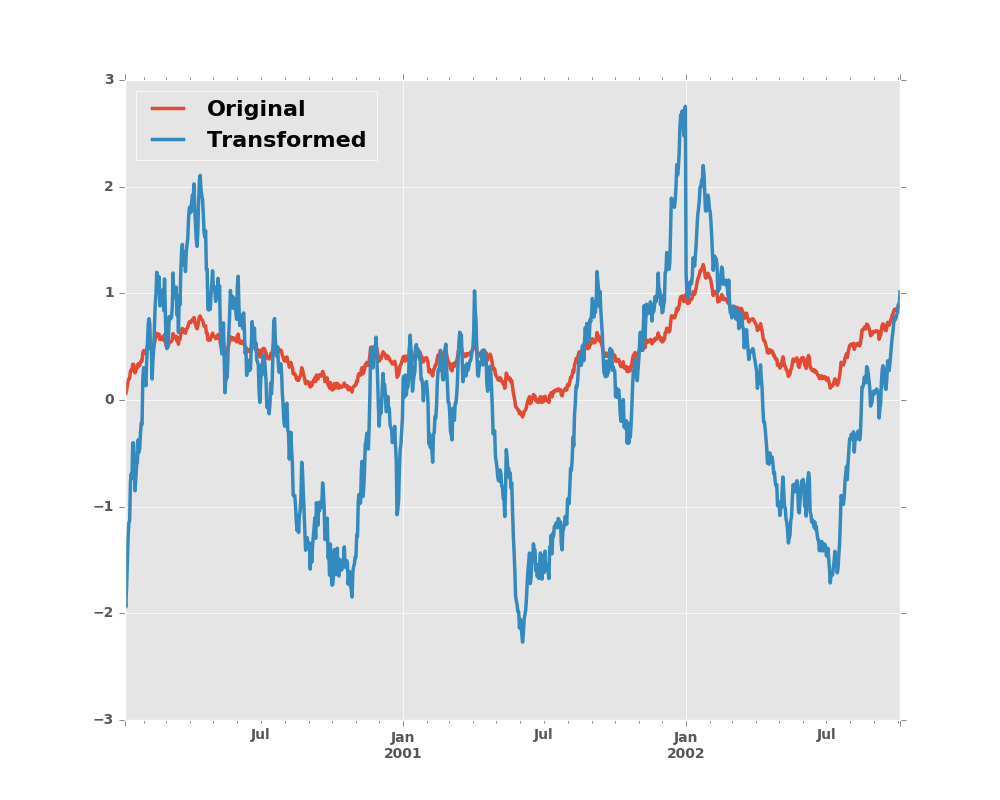
We can also visually compare the original and transformed data sets.
In [15]: compare = pd.DataFrame({'Original': ts, 'Transformed': transformed})
In [16]: compare.plot()
Out[16]: <matplotlib.axes._subplots.AxesSubplot at 0x2b35be9a4a90>
Another common data transform is to replace missing data with the group mean.
In [17]: data_df
Out[17]:
A B C
0 -0.786641 -0.393609 -0.171092
1 1.786173 -0.575189 0.820750
2 -1.256530 -0.642246 0.236892
3 -0.466867 -1.382653 -0.369422
4 NaN -0.851236 1.058006
5 -0.080372 -1.268121 1.561967
6 0.816983 1.965656 -1.169408
.. ... ... ...
993 NaN -0.575634 -2.332083
994 0.225519 0.347968 -0.517897
995 0.945827 0.190394 0.845686
996 0.607557 0.722559 0.737809
997 0.024958 0.553376 0.477220
998 0.647037 -0.397862 -0.022409
999 0.929380 0.232026 -1.296769
[1000 rows x 3 columns]
In [18]: countries = np.array(['US', 'UK', 'GR', 'JP'])
In [19]: key = countries[np.random.randint(0, 4, 1000)]
In [20]: grouped = data_df.groupby(key)
# Non-NA count in each group
In [21]: grouped.count()
Out[21]:
A B C
GR 208 221 189
JP 235 251 215
UK 235 239 217
US 230 239 202
In [22]: f = lambda x: x.fillna(x.mean())
In [23]: transformed = grouped.transform(f)
We can verify that the group means have not changed in the transformed data and that the transformed data contains no NAs.
In [24]: grouped_trans = transformed.groupby(key)
In [25]: grouped.mean() # original group means
Out[25]:
A B C
GR 0.032022 0.037422 0.007687
JP -0.049426 -0.008097 -0.092958
UK 0.072942 0.059592 0.176494
US -0.016772 -0.075033 -0.086060
In [26]: grouped_trans.mean() # transformation did not change group means
Out[26]:
A B C
GR 0.032022 0.037422 0.007687
JP -0.049426 -0.008097 -0.092958
UK 0.072942 0.059592 0.176494
US -0.016772 -0.075033 -0.086060
In [27]: grouped.count() # original has some missing data points
Out[27]:
A B C
GR 208 221 189
JP 235 251 215
UK 235 239 217
US 230 239 202
In [28]: grouped_trans.count() # counts after transformation
Out[28]:
A B C
GR 234 234 234
JP 261 261 261
UK 255 255 255
US 250 250 250
In [29]: grouped_trans.size() # Verify non-NA count equals group size
Out[29]:
GR 234
JP 261
UK 255
US 250
dtype: int64
Note
Some functions when applied to a groupby object will automatically transform the input, returning
an object of the same shape as the original. Passing as_index=False will not affect these transformation methods.
For example: fillna, ffill, bfill, shift.
In [30]: grouped.ffill()
Out[30]:
A B C
0 -0.786641 -0.393609 -0.171092
1 1.786173 -0.575189 0.820750
2 -1.256530 -0.642246 0.236892
3 -0.466867 -1.382653 -0.369422
4 -0.786641 -0.851236 1.058006
5 -0.080372 -1.268121 1.561967
6 0.816983 1.965656 -1.169408
.. ... ... ...
993 -0.661776 -0.575634 -2.332083
994 0.225519 0.347968 -0.517897
995 0.945827 0.190394 0.845686
996 0.607557 0.722559 0.737809
997 0.024958 0.553376 0.477220
998 0.647037 -0.397862 -0.022409
999 0.929380 0.232026 -1.296769
[1000 rows x 3 columns]